Our Research
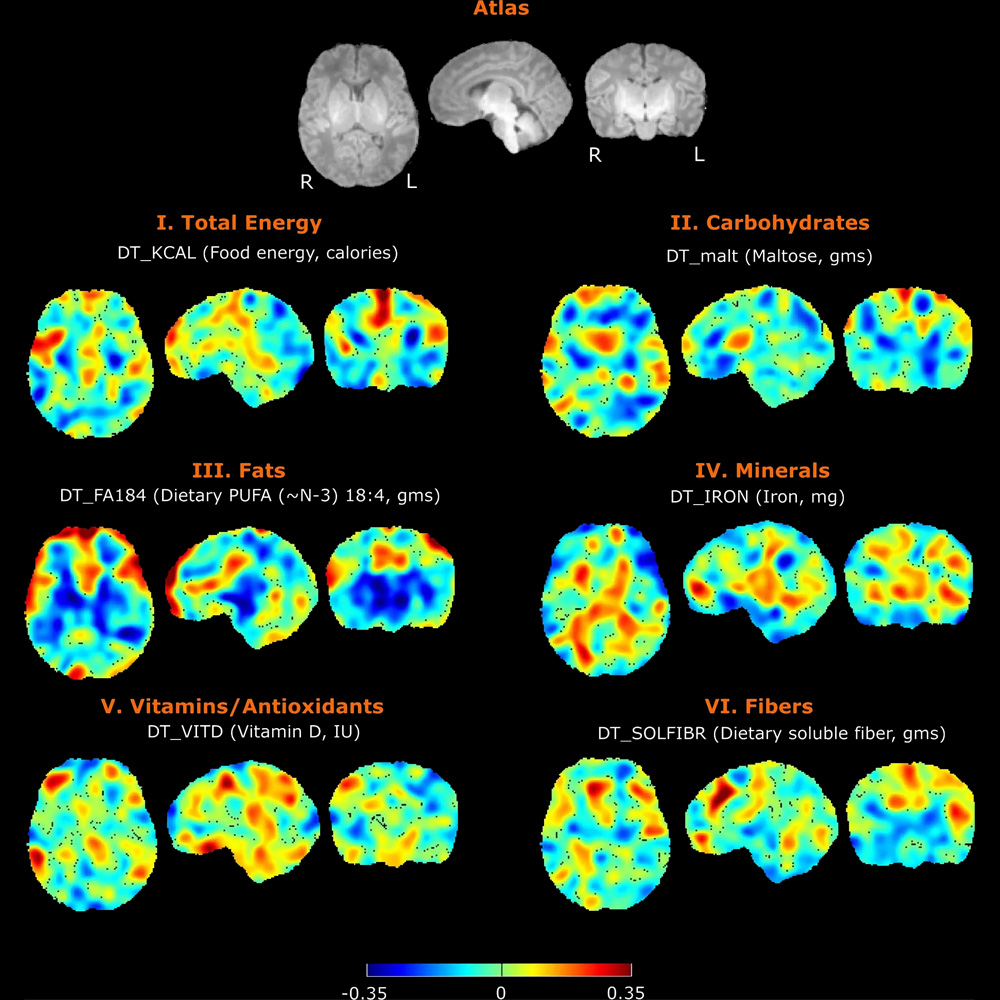
Nutrition and Infant Neurodevelopment
Nutrition has profound impact on neurodevelopment, but the mechanisms are only partly understood. In collaboration with other researchers at the Fetal-Neonatal Neuroimaging Developmental Science Center, we are studying the connections between maternal diet, breastmilk contents, infant brain development and child neurodevelopment. We also have a pilot study on the trajectory of infant develop the important skill of oral feeding with the goal of improving diagnosis and personalized care through quantitative EMG assessment of infant feeding, advanced computational analytics, and identifying biomarkers of neonatal outcomes.
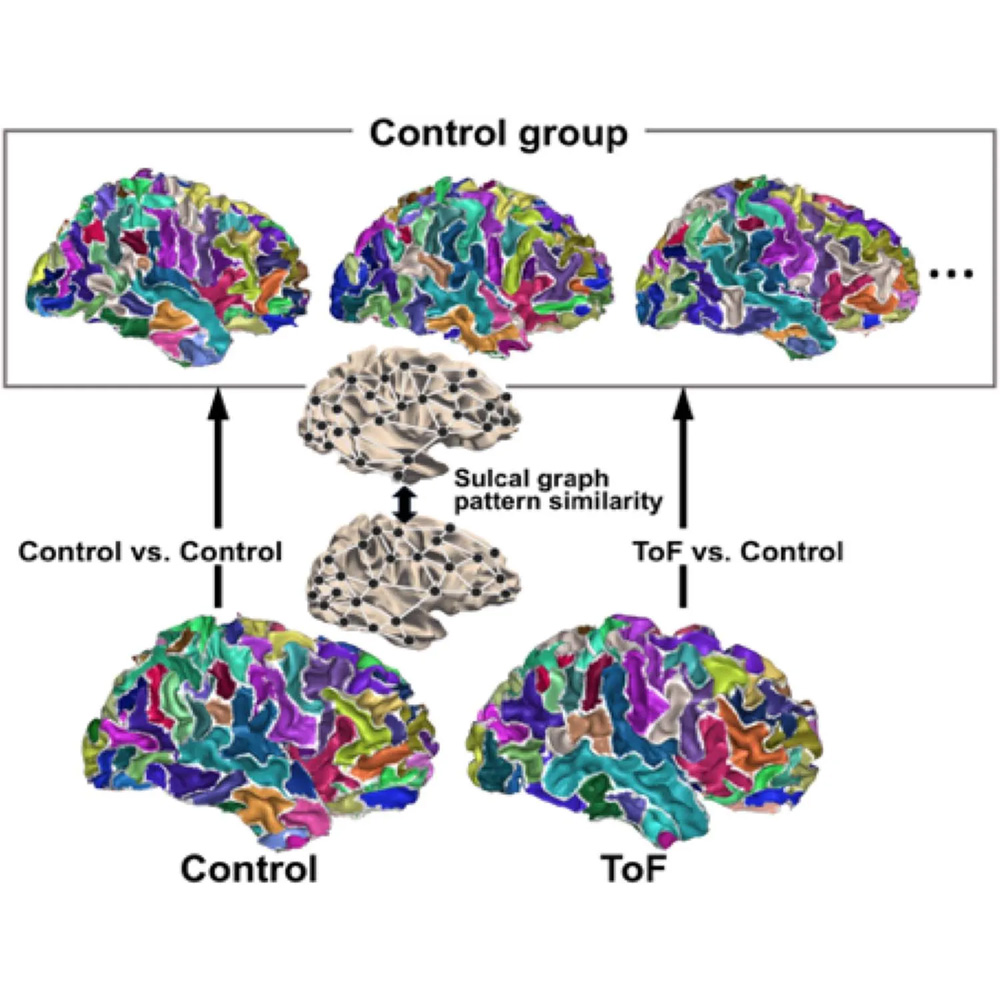
Modifiers of Neurodevelopment among Patients with Congenital Heart Disease
Congenital heart disease (CHD) is the most common severe malformation. As improvements in medical and surgical management have led to increased survival, patients with congenital heart disease face additional lifelong health risks. Neurodevelopmental delay or impairment is the most common extracardiac complication of CHD. To better understand the mechanisms of neurodevelopmental risk in patients with CHD, we have recently participated a clinical trial that collected genetic, clinical, and neuropsychological testing data. Ongoing projects include further analysis of that trial data, and local pilot studies.
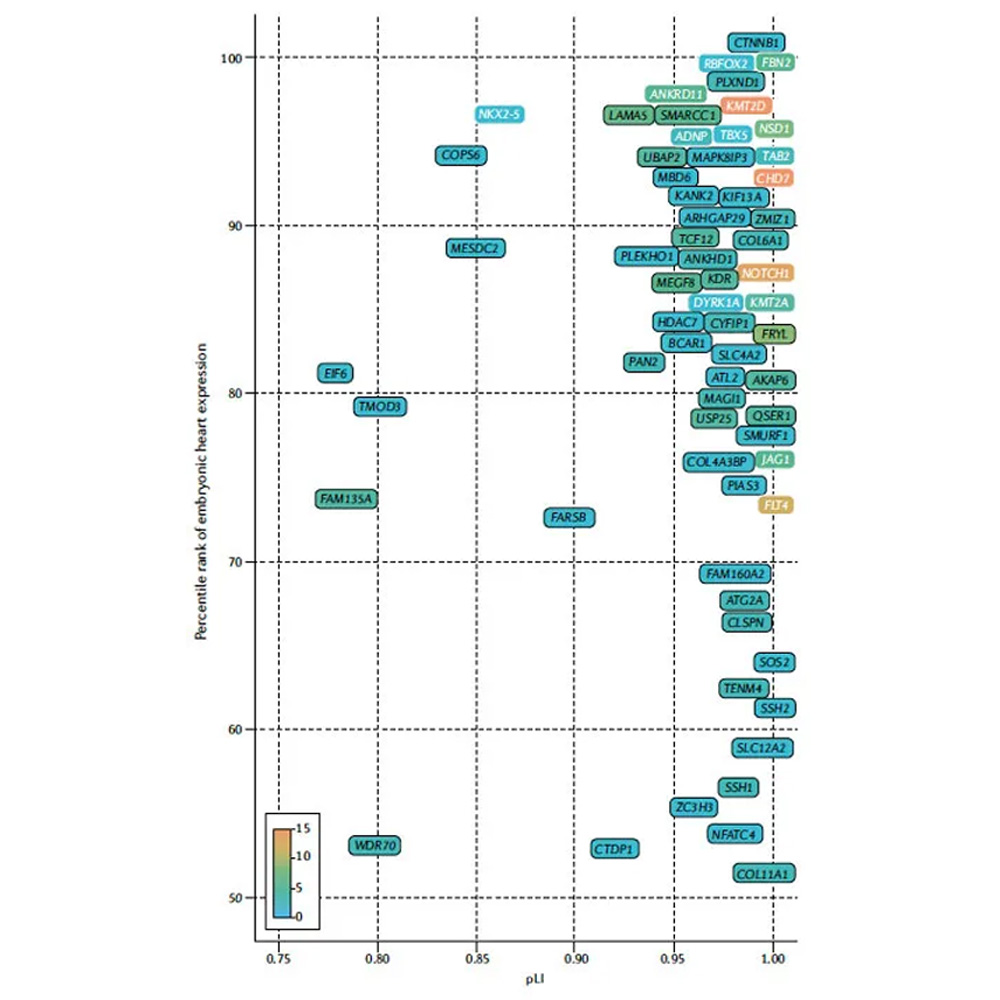
Gene Discovery in Congenital Heart Disease
We study the genetics of congenital heart disease with the goal of improving diagnosis and personalized care through gene discovery, functional analysis of patient variants, and identifying biomarkers of neonatal outcomes. Approaches include computational biology projects, cell culture projects, and multi-omic analysis of patient samples.

